Neuroscience
Advancing experimental and theoretical neuroscience research
Our mission is to support the work of neuroscientists to fully reveal the structure of the brain, and inform groundbreaking new treatment options for mental illnesses.
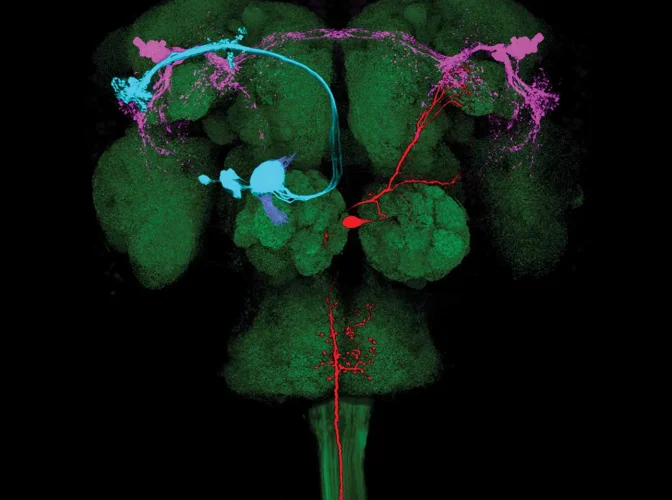
Our motivation
Our focus is on fundamental research, with a portfolio that spans genes to behaviour, investing in science that is not driven by one single disorder or the pursuit of a cure for mental illness. Our challenge is in identifying, observing and ultimately understanding the circuits that generate behaviour. The knowledge accumulated through these studies will underpin many of the future breakthroughs in the treatment of functional neurological disorder.
In addition to funding research that will advance knowledge in experimental and theoretical neuroscience, we support a range of related activities that help develop the wider neuroscience sector. For example, we support conference series for sharing the latest data and networking, as well as advanced training workshops to disseminate the latest research methods as well as nurture the research leaders of the future.
Creating centres of excellence
Developing some of the world’s leading institutes for neuroscience research
Creating an experimental research portfolio
Supporting cutting-edge fundamental research in neuroscience
Supporting neuroscience theoreticians
Fostering collaborations with other institutes around the world
Developing the neuroscience sector
Investing in strategic programmes to develop the sector
Forming strategic partnerships
Collaborating with like-minded organisations to support neuroscience research

Background & objectives
Gatsby’s pioneering investments in neuroscience began in the 1990s in the area of theoretical neuroscience with the establishment of the Gatsby Computational Neuroscience Unit at University College London (UCL).
In 2007, the Trustees decided to expand the investment in this exciting field, and embarked on an ambitious programme of funding experimental research projects in the area of neural circuits and behaviour. Activity was delivered via supporting consortia around the world and building a major new research institute, the Sainsbury Wellcome Centre for Neural Circuits and Behaviour, at UCL.
The Centre opened in 2016, and hosts a diverse group of experts from different disciplines including computation, physics, chemistry and engineering. The shared mission of these individuals is to progress our understanding of how behaviour is generated from the complex patterns and properties of neurons.
Funding opportunities
We are assisted in our neuroscience programmes by Dr Sarah Caddick, CEO of Thalamic Ltd.
Our funding is directed and proactive. We do not accept unsolicited applications; however we are always keen to establish strong links with other funders, programmes and centres that share our goal of advancing neuroscience research, and complement our current investments.